In the previous posts of this series on genetics and racism, I talked about two recent academic disputes over human races. With this post I hope to give a wider overview of what biology has to say about species, breeds and races.
Darwin’s pigeons
 Moderngenetics was born in 1900 with the re-discovery ofMendel's laws. Since theNeolithic Revolution, genetics had been an empirical art. Our ancestors isolated most of the breeds of animals and plants that we know today, i.e.groups that carry a trait of interest to the next generation when crossed together (for instanceChihuahuas are small dogs and Great Dane are large dogs).
Moderngenetics was born in 1900 with the re-discovery ofMendel's laws. Since theNeolithic Revolution, genetics had been an empirical art. Our ancestors isolated most of the breeds of animals and plants that we know today, i.e.groups that carry a trait of interest to the next generation when crossed together (for instanceChihuahuas are small dogs and Great Dane are large dogs).
But over the generations, pedigrees got lost in the myst of time and the overwhelming differences between some breeds of the same species raised the question whether they share the same natural origin. Before Darwin, it was difficult to imagine that the Chihuahuas and the Great Dane would have a common ancestor, and the theory went that breeds actually came from different species. This is actually one of the first questions tackled by Darwin in The Origin of Species. In the following passage, he exposes his conclusions after a hybrid cross between different breeds of pigeons.
Great as the differences are between the breeds of pigeons, I am fully convinced that the common opinion of naturalists is correct, namely, that all have descended from the rock-pigeon (Columba livia), (...) I crossed some uniformly white fantails with some uniformly black barbs, and they produced mottled brown and black birds; these I again crossed together, and one grandchild of the pure white fantail and the pure black barb was of as beautiful a blue colour, with the white rump, double black wing-bar, and barred and white-edged tail-feathers, as any wild rock pigeon!
Even if Darwin could not figure out the genetics of pigeon colour, he understood that the fantail and the barb were different exploits of the variability of the rock pigeon. And in fact, domestic breeds are not part of a natural, pre-established order, they are human artifacts.
Bits and species
In the quote above, Darwin refers to the rock pigeon as Columba livia. These Latin and Greek names were introduced as a part of the great endeavor of the Swedish naturalistCarl Linnaeus to classify all living organisms. Taxonomy, the classification of living organisms, rests on the tacit assumption that species form essentially distinct groups. However, speciation (the appearance of new species) is an ongoing process. Between the time when a species does not exist and the time it comes into existence, there is a grey zone where the population is somewhere in between one and two species.
What is the point of classifying organisms if the classes keep changing? As traumatic as the theory of Evolution was for taxonomy, it gave it a more meaningful and noble aim, namely to recapitulate the tree of life. In other words, it is understood that whenever possible, taxonomy must coincide with phylogenetics. This principle in mind, Ernst Mayr proposed the definition of species that is the most commonly accepted today.
Groups of actually or potentially interbreeding natural populations, which are reproductively isolated from other such groups.
In other words, two individuals are from the same species if they can have fertile descendants together. Why is that a better criterion than having the same number of legs, or the same hair color? Reproductively isolated populations have no way of exchanging their genes, so their characters will diverge as time goes by. Conversely, if a character appears in a group of inter-fertile individuals, it can spread by descent in the population, which remains more homogenous over time.
This definition is not without difficulties, such as the existence of ring species (groups of species that can be inbred with their geographical neighbors but not with the neighbors of their neighbors) or populations without sexual reproduction. Yet, it captures an essential aspect of the genetic dynamics of natural populations.
For races though, things get more difficult. Again according to Ernst Mayr
a subspecies is a geographic race that is sufficiently different taxonomically to be worthy of a separate name.
So here the purpose is to know what we are talking about and keep the collections of stuffed animals well ordered in the museums. This definition admirably illustrates the difficulty of the concept of race. Species can show geographical variations, but is it a property of the geography or a property of the population?
Ceci n’est pas une race
 Finding objective criteria to delineate races is still an ongoing debate among biologists. One of them in particular,genetic diversity — which was already touched upon in the first post of this series — is often put forward as a natural choice. Is it reasonable to qualify as different races two populations that are sufficiently different from the point of view of genetic variation? Since genetic differences keep increasing between species, it is tempting to use it as a measure of ongoing speciation.
Finding objective criteria to delineate races is still an ongoing debate among biologists. One of them in particular,genetic diversity — which was already touched upon in the first post of this series — is often put forward as a natural choice. Is it reasonable to qualify as different races two populations that are sufficiently different from the point of view of genetic variation? Since genetic differences keep increasing between species, it is tempting to use it as a measure of ongoing speciation.
An important issue, mentioned by Johnathan Kaplan in the comments of thefirst post of this series, is that the way of measuring genetic diversity matters. There is a cornucopia of indicators out there, and the attitude is often to consider them equivalent. In reality they measure one of three different quantities, namelydiversity (allele richness),differentiation (allelic distance) or heterozygosity.
Most studies of the variability of human races use indicators of the third type, which do not meet the expectations of conservation biologists. Kaplan and Winther[1] give the example (due to Lou Jost) of two sub-populations, each with ten equally frequent alleles, and such that each allele is in only one population. Using the same measure asLewontin, we would conclude that 77% of the variability is within sub-populations (the calculation is detailed in the technical section below). Still, we would lose 50% of the alleles should one of these sub-populations disappear. The choice of heterozygosity is somewhat arbitrary and uninformative regarding diversity and differentiation. However, the choice of one or the other will lead to the conclusion that the differences between populations are large or negligible, giving support for or against the existence of races.

But genetic diversity is not universally recognized as a good criterion. Genetic differences do not need to be large between different species because speciation does not need to be a gradual phenomenon. Mosquitoes Anopheles gambiae actually consist of two reproductively isolated populations that constitute two species separated less than 10,000 years ago[2]. Only three regions representing ~ 1% of the genome are fixed between the two species. The alleles of the remaining 99% are the same in both species, but they occur with different frequencies. In this example, a definition of race based on genetic diversity would lead to the paradoxical conclusion that Anopheles gambiaeconsists of two species, but of a single race.
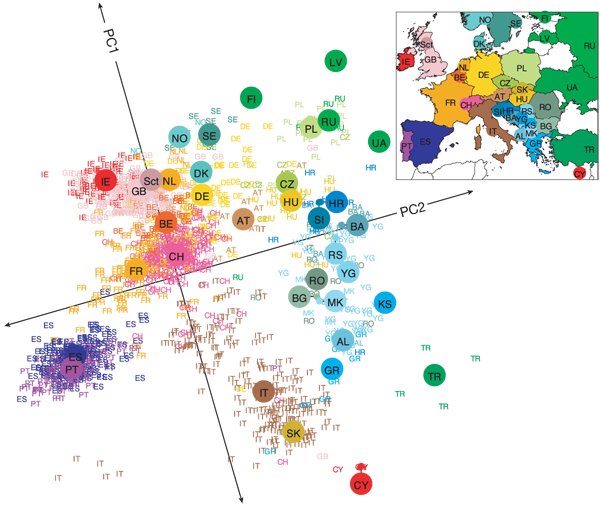
Genetic clustering of the Europeans by Principal Component Analysis shows that genetic fingerprints correlate with geographic origin of the individuals. Reproduced from reference [3] with permission from John Novembre.
Even when genetic differences are small, human races can be discriminated byclustering analysis, provided enough loci are considered — this is the argument put forward by Anthony W.F. Edwards in Lewontin's fallacy (see the first post of this series for the details). Population history and local inbreeding leave marks on the genome. In Europe for instance, genetic markers can reveal your origin within a 500 km radius[3](see figure above).
If genetics can discriminate human populations, why don't we use them as races, you may ask? And I would return the question, why would you use populations as races?The term race is loaded with so many connotations that it cannot be used in a purely scientific context. It has never been, it probably never will. It does not belong to genetics to define human races.



 and validity of these scientific findings. Although historically there have been reasons for distrust of science, sociologists need not assume that science actively seeks to create or maintain racial hierarchy. In science there is truth and means for understanding our world. The scientific community and the social science community need to work together to make their work accessible and understandable for each other as well as the general public. Proper analysis of the complex ways in which society and genetics interact requires interdisciplinary discussion, open-mindedness, and most importantly the building of bridges that will allow all to cross into a place of understanding.
and validity of these scientific findings. Although historically there have been reasons for distrust of science, sociologists need not assume that science actively seeks to create or maintain racial hierarchy. In science there is truth and means for understanding our world. The scientific community and the social science community need to work together to make their work accessible and understandable for each other as well as the general public. Proper analysis of the complex ways in which society and genetics interact requires interdisciplinary discussion, open-mindedness, and most importantly the building of bridges that will allow all to cross into a place of understanding.



